Read more about Direct PCR of Crude Samples without Template Purification, highQu PCR & qPCR Reagents Performance Excellence etc.
Genome-wide bulk sequencing is widely used to assess DNA methylation (DNAm). However, methylation signals obtained from bulk sequencing are averaged out, concealing the cell-to-cell differences in methylation signatures involved in health and disease. Single-cell methods such as bisulfite sequencing were developed to reveal these signatures and offer insights into cellular heterogeneity and rare cell types. However, genome-wide DNAm profiling at single-cell resolution has drawbacks such as sparse data and limited genomic coverage even for deeply sequenced samples.
To address this gap, the Renée Beekman Lab and the Lars Velten Lab of the Centre for Genomic Regulation (CRG) in Barcelona teamed up to develop a novel high-throughput, high-confidence targeted bisulfite-free method for analyzing DNAm in single cells, now published in Genome Biology.
In this webinar, authors Michael Scherer, Ph.D. and Agostina Bianchi of CRG will present the Single-Cell Targeted Analysis of the Methylome (scTAM-seq) protocol which profiles 650 biologically relevant CpGs in up to 10,000 cells using the Tapestri platform. They will also share how they combined DNAm (epigenetics), somatic mutations (genotype), and surface marker expression (phenotype) to gain a rich picture of clonal architecture and dynamics across B cell differentiation in blood and bone marrow.
scTAM-seq is a high-throughput, targeted restriction enzyme-based approach for measuring DNA methylation states at single-cell resolution. The method involves
the use of methylation-sensitive restriction enzymes (MSRE’s) to selectively cut unmethylated recognition sites, while leaving methylated sites intact.
scTAM-seq is a powerful method for investigating DNA methylation dynamics at single-cell and single-nucleotide resolutions
scTAM-seq enables DNA methylation assessment with high accuracy, achieving a false-positive rate lower than 0.2% combined with a false negative rate as low as 7%
scTAM-seq can be combined with somatic mutation and cell surface expression analysis to enable a much richer characterization of tumor heterogeneity
Tapestri single-cell multiomics analysis can transform how you understand the complexities of multiple myeloma and its precursor stages by integrating genomic, proteomic, and clonotypic subclonal assessment with single-cell resolution, all within a single assay.
The assay includes coverage of key driver and resistance genes, genome-wide copy number variations (CNVs), V(D)J clonotype, and cell lineage & immunotherapy markers.
As the only solution to integrate genotypic and immunophenotypic assessment, the scMRD AML Multiomics Assay targets 40 genes for single-cell DNA sequencing based on current international AML MRD guidelines, such as European LeukemiaNet, and 17-plex antibody-oligonucleotide conjugate (AOC) panel curated for key biomarkers associated with AML MRD.
Through a seamlessly integrated workflow, the assay allows clinician-researchers to:
Acute myeloid leukemia (AML) measurable residual disease (MRD) is difficult to detect accurately, and current methods such as flow cytometry (FCM) and bulk next-generation sequencing (NGS) are commonly challenged by both false-positive and false-negative results. Even when these single-metric assays agree on
an MRD result, too often they are still discordant with clinical outcome.
The new scMRD assay from MissionBio allows genotypic and immunophenotypic analysis at clonal level with single cell resolution for precision medicine.
Tapestri® Platform powers Nature Medicine publication that identified rare subclones associated with aggressive cancer progression
The Ionic system is so different, its advantages are most readily understood in contrast with conventional nucleic acid extraction and
purification methods:
Higher nucleic acid yields; No sample loss associated with binding nucleic acids to, or stripping from, fixed surfaces
Simple workflows with fully automated separations; No columns or beads and no repetitive washing
Reduced nucleic acid fragmentation; No harsh high-salt buffers or organic solvent
Tapestri® Platform powers Nature Medicine publication that identified rare subclones associated with aggressive cancer progression
In new findings from the lab of Elías Campo, Director of the August Pi i Sunyer Biomedical Research Institute (IDIBAPS) in Barcelona, Spain, which used single-cell DNA sequencing to identify genetic drivers of Richter’s syndrome, the unpredictable and often deadly progression from chronic lymphocytic leukemia (CLL) to an aggressive B cell lymphoma.
The new study, published in the journal Nature Medicine, was the latest to demonstrate clinically relevant insights using Mission Bio’s Tapestri® Platform, highlighting rare Richter’s Syndrome subclones that lie dormant for years and could be targeted therapeutically to prevent disease progression by uncovering mutations at the individual cell level.
A major challenge to combating cancer is the tumor’s inherent heterogeneity. Single-cell DNA sequencing allows you to unmask the underlying genetic diversity across cell populations and gain insights into the clonal architecture, mutation co-occurrence, and rare mutations driving tumor progression and therapy resistance.
Tapestri Single-cell DNA Breast Cancer Research Panel V2
Advance your understanding of the genetic heterogeneity underpinning breast cancer by targeting 32 hotspot genes and 30 copy number variants and chromosome arm aneuploidies for single-cell sequencing. Carefully curated based on evidence from more than 15 databases such as COSMIC and reviewed by leading researchers of breast cancer, this panel is designed to cover some of the most commonly mutated genes associated with breast cancer, including BRCA1 and BRCA2
Measurable residual disease (MRD) detection using next-generation sequencing (NGS) has allowed for a sensitive evaluation of a patient’s relapse risk, but several challenges remain unresolved. Bulk NGS alone provides limited sensitivity and specificity to deconstruct the complex heterogeneity/somatic mosaicism driving disease transformation or to distinguish between leukemic clones and clonal hematopoiesis (CH). It also lacks information about immunophenotypic heterogeneity, typically inferred by flow cytometry, rendering an incomplete picture.
Mission Bio’s Tapestri Platform is the first single-cell multi-omics approach integrating molecular (mutational) profiling and immunophenotyping in a single assay to allow for scMRD detection with unparalleled resolution, sensitivity and specificity, ultimately improving identification of residual disease and potential therapeutic strategies
Tapestri Platform resolves several challenges in MRD testing:
Unmask clonal architecture at diagnosis and during treatment and detect rare and subclonal variants driving the disease transformation
Characterize changes in clonal diversity over time, providing valuable information about tumor evolution and its correlation with relapse
Enable unambiguous distinction between leukemia driving clones and CH, reducing false positives
Integrate single-cell genotypic and immunophenotypic information for a comprehensive MRD assessment
Celemics Whole Exome Sequencing Service includes
Celemics Whole Exome is a comprehensive whole exome panel that covers the target
regions of major WES panels in the market.
With the target size of 37 Mb, the panel does not compromise performance in terms of coverage and uniformity, enabling highly efficient and cost-effective sequencing of the whole human exome.
Celemics panels also perform well against hard-to-capture regions such as GC-rich regions.
Regarding spike-in options, we also have the ability to customize our WES panel according to your needs by including mitochondrial or intronic regions at your request.
The panel is also supported by Celemics Analysis Service (CAS),
an end-to-end bioinformatics solution.
Detects ctDNA for colorectal cancer, breast cancer, and lung cancer
Assess 16 key genes for colorectal cancer, 27 for breast cancer, 28 for lung cancer
Highly optimized panel for clinical testing with exceptional accuracy
Complete validated panel performance conducted with patient samples
Respiratory viruses including SARS-CoV-2 were detected in air samples using the Celemics target enrichment technology.
The CRV panel was used for hybridization capture of multiple viruses at highly sensitive capture performance and compared against RT-PCR.
Different variant types such as SNV´s, indels and rearrangements were successfully detected.
The Celemics CRV panel includes a standalone, easy-to-use bioinformatics software.

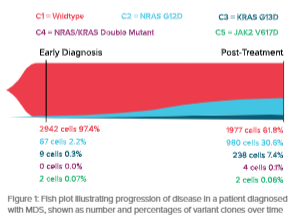
The Tapestri Single-Cell DNA AML Panel was used to comprehensively analyze the mutational landscape of MDS patient samples. Whether testing for diagnosis or disease monitoring, these results showed high sensitivity, clonal resolution, and concordance of variant allele frequency (VAF) between single-cell and bulk next generation sequencing (NGS) data.
Additionally, clonal variant co-occurrence was resolved, allowing for clonal lineage reconstruction, and a more comprehensive picture of the patients’ disease
Takeaways:
Recurrent somatic mutations in MDS overlap with the Tapestri Single-Cell DNA AML Panel
As few as 2 cells were detected at a sensitivity of 0.06%
Single-cell genomic analysis reveals mutational co-occurrence and clonal evolution.
The Celemics G-Mendeliome Disease-Specific Panels are designed to analyse genes in various inherited diseases assiciated with:
AML, ALL, Multiple Myeloma, Lymphoma, Hearing loss, Hereditary cancer syndrome, Arrhythmia,Cardiomyopathy, Muscular dystrophy, Charcot-Marie-Tooth, Spastic paraplegia, Myopathy, Ataxia, Epilepsy, Parkinson, Alzheimer, Dementia, and Dystonia
The Celemics G-Mendeliome CES Panel allows comprehensive genomic profiling of a variety genetic disease and covers up to 7513 genes across clinically significant regions not covered by other panels. The G-Mendeliome CES Panel has been optimised in collaboration with high level clinical NGS service providers. It is a cost-effective options to WES.
riboPOOls’ unique feature is their high complexity pooling approach based on our Pack Hunter design algorithm.
The high complexity of riboPOOLs ensures optimal and maximum rRNA coverage, allowing for efficient and cost-effective rRNA removal of any species or abundant RNA.
riboPOOLs are available for single species (e.g., Homo sapiens, Drosophila melanogaster, etc.) and multiple species.
Benefits of riboPOOLs
rRNA depletion for any species
High complexity probes for efficient, reliable rRNA removal
Cost-effective
Wide RNA-input range (10 ng – 3 ug)
riboPOOLs for highly degraded samples (FFPE)
Special solution for ribosome profiling
Single cell multiomics assays add to sample complexity. The necessity of removing excess assay reagents to reduce background noise is critical to achieve high quality data from cell suspensions or nuclei samples.
By using Laminar Mini, scientists can gently remove unbound antibodies, debris and dead cells from multiomics samples. The wash workflow saves lot of time and retains precious, small populations.
The Celemics Comprehensive Respiratory Virus Panel (CRV Panel) was developed to detect and sequence respiratory disease-causing viruses in humans using the NCBI RefSeq database as its foundation.
It allows for the Whole Genome Sequencing of SARS-CoV-2 and all its relevant mutations and variants, and enables simultaneous testing of 9 different virus types and its 39 strains of clinically significant and prevalent respiratory viruses.
Using the Laminar Wash system saves time with fewer manual manipulations. Gently washing cells without unnatural centrifugal forces maintains cell integrity and viability, while ensuring gene or protein expression changes are biological, instead of due to workflow.
Laminar Wash technology has been added as part of the official TotalSeq™ Protocol. It simplifies the processing of samples with low cell counts, enabling high-throughput.
Total RNA extracted from FFPE tissue (Formalin-fixed, paraffin-embedded tissues) are heavily degraded with RIN values below 2. Due to their strong fragmentation ribosomal RNAs are difficult to remove before RNA sequencing (RNA-Seq). With a gapless probe coverage of the entire rRNA sequence, siTOOLs Biotech developed an efficient rRNA depletion tool for FFPE and other strongly degraded RNA samples called FFPE riboPOOL.

The Celemics Comprehensive Respiratory Virus Panel (CRVP) is able to yield affirmative results even if a clinical specimen is of low quality, and can also test for double pandemic and co-infection possibilities.
Designed to identify, detect and sequence each pathogen and any present mutations in a single reaction, it reveals information on the epidemiology and pathogenesis of viruses.
As multiple respiratory viruses can be detected at once, differentiation of Covid-19 against other infections such as seasonal flu, helps avoiding unnecessary quarantines and saves time and money.
The Virus Verifier stand-alone bioinformatics package is included with the kit. It provides a full vire of sequencing data, virus distribution, coverage per detected strain and the 1X coverage level for those strains.
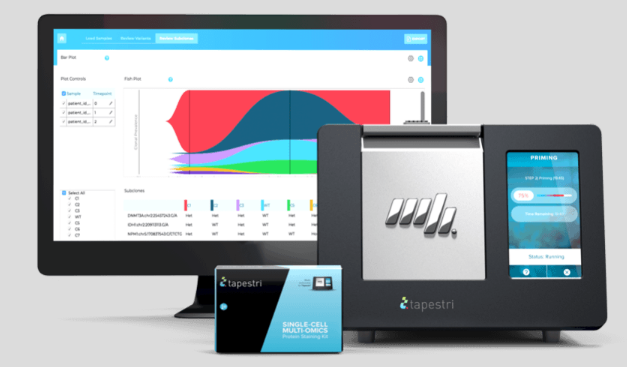
The Tapestri Platform analyzes genotype and phenotype simultaneously from the same single cells, letting you reveal clonal heterogeneity and target comprehensive biomarkers that help stratify patients more precisely, signal resistance as it begins, and predict relapse.
Check our latest Application Note on the study of single cell genomic variants and protein expression correlations in AML patients.
The h/m/r riboPOOLs offer rRNA depletion from human, mouse or rat samples at high or low RNA quality.
Target cytoplasmic rRNA (28S, 18S, 5.8S & 5S rRNA) & mitochondrial rRNA.
Enzyme-free, fast & easy hybridization based workflow.
Any reaction sizes (6, 12, 24, 96, …).
Wide input range (10 ng – 3 μg).
Combination with other ready-made riboPOOLs possible, such as Globin mRNA.
Low quality samples/FFPE:
riboPOOL human/mouse/rat degraded RNA kits
For RIN<7
100% coverage of ribosomal RNA
87-98% rRNA depletion efficiency
High quality samples:
riboPOOL human/mouse/rat kits
RIN>7
97-98% rRNA depletion efficiency
Rib-seq/Ribosome profiling:
riboPOOL ribo-seq human/mouse/rat kitsRibosome Profiling (30-mer contaminants)
100% coverage of rRNA
Adjusted to abundancy of rRNA contaminants
53%-75% rRNA depletion efficiency
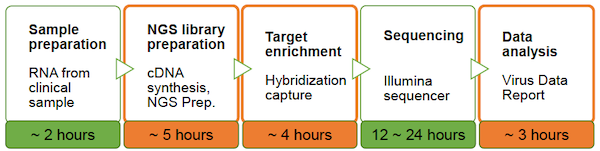
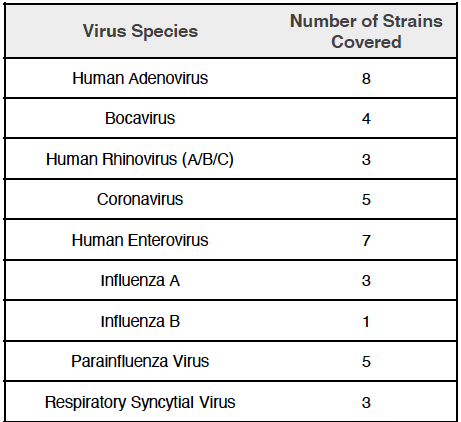

Covering a wide range of respiratory pathogens: 39 different strains from 9 virus types, including SARS-CoV-2, Influenza A/B, RSV A/B and more, the CRVP panel gives a broader picture for not only detection of pathogens but also to study co-infection.
A dedicated bioinformatics data analysis package is included.
Pathogen detection is made at high sensitivity and consensus sequence generation rate down to 100 copies. More than 100X coverage of 99% of viral genome.
The panel was tested using low-quality clinical samples.
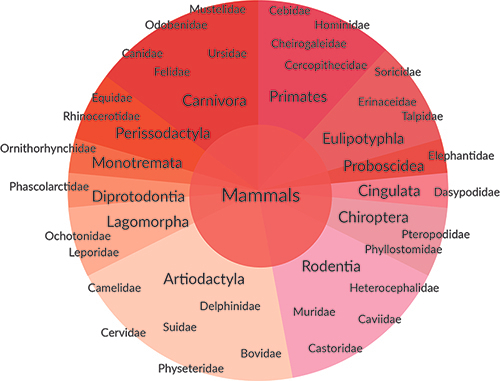
The high complexity riboPOOLs offer efficient removal from a wide range of species.
Multiple-species Pan-riboPOOLs for depletion from similar species:
Pan-Mammal
Pan-Plant
Pan-Bird
Pan-Prokaryote
Pan-Bacteria
Pan-Actinobacteria
Pan-Archaea
Pan-Fungi
Multiple species Combination riboPOOLs for depletion from diverse species groups:
Select from existing riboPOOLs or Pan-riboPOOLs and combine in a ratio reflecting the amount of rRNA from the different species in your sample
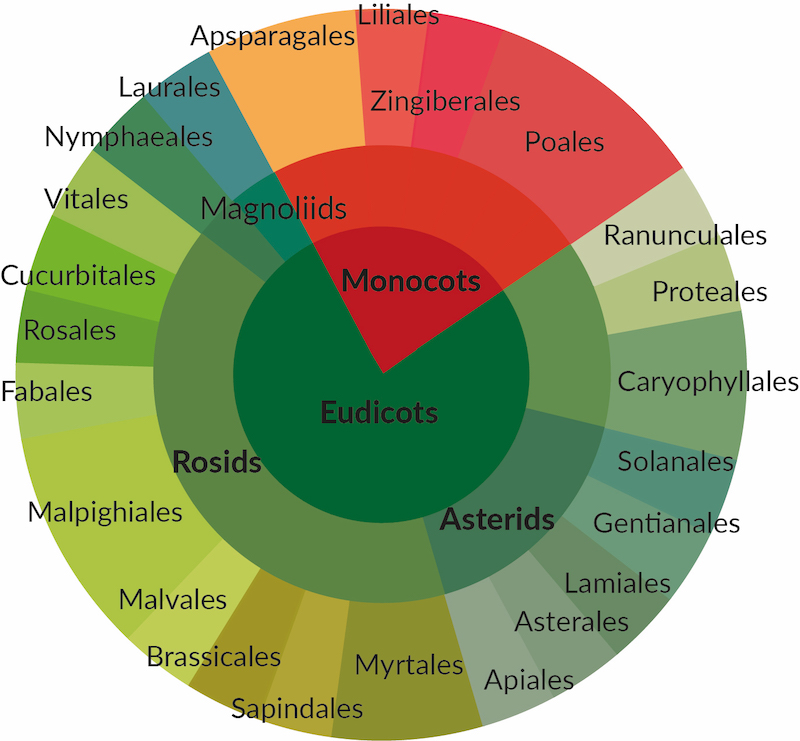
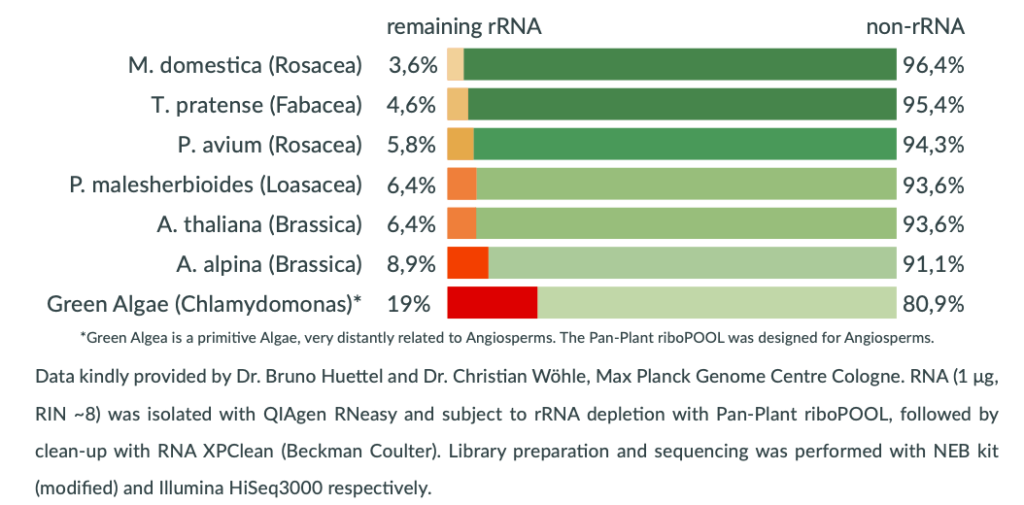
Efficient rRNA depletion
Broad coverage of flowering plants
For leaf, seed and root tissue
Targets 28S, 18S, 5.8S & 5S rRNA
Targets mitochondrial rRNA
Targets plastid rRNA
The high abundance of ribosomal RNA (rRNA) limits detection efficiency of relevant RNAs of the transcriptome by RNA-Seq. The Pan-Plant riboPOOL offers a universal solution to selectively deplete cytoplasmic (28S, 18S, 5.8S & 5S), plastid and mitochondrial rRNA of flowering plants in leaf, seed and root tissue.
The Pan-Plant riboPOOL efficiently depleted rRNA (> 91%) when tested on six species of Angiosperms from genera Fabacea, Loasacea, Brassicae and Rosaceae. Remarkably, a species from the primitive Green Algae phyla when tested showed > 80% rRNA depletion efficiency, suggesting that the Pan-Plant riboPOOL may be successfully applied to other phyla aside from Angiosperms.
Celemics PharmacoScreen panel encompasses 115 genes related to Pharmacokinetics and Pharmacodynamics
1. Phase I/II drug-metabolizing enzyme genes (drug metabolism)
2. ABC & SLC family transporter genes (drug effect)
3. Pharmacodynamics genes (drug biochemical and physiological mechanism)
4.Modifier genes (drug ADME enhancement).
The panels feature repetitively optimized probes to capture up to 115 PK/PD-related genes with high coverage (99.6%) and accuracy (99.9%). Sequencing of a Korean cohort (n=376) with the panels enabled profiling of actionable variants as well as rare variants of unknown functional consequences.

Tapestri workflow provides a great possibility in single-cell DNA sequencing. SNVs, CNVs and protein expression can be studied simultaneously at single cell level.
Panels available for hematology, solid tumour profiling, genome editing, SNVs, SNVs+CNVs, SNVs+CNVs+protein

In a collaboration the Korean Centre for Disease Control, Celemics have developed BT-Seq for fast and accurate whole genomce sequencing of small genomes. The technology was used to determine the sequence of Covid-19, isolated in South Korea.
BT-Seq is based on a unique molecular barcode and enzyme technology. Together with a proprietary bioinformatics algorithm, BT-Seq delivers data in short time at the same accuracy as Sanger sequencing.
The technology is currently available as a service and collaboration. A kit will be launched during 2020.
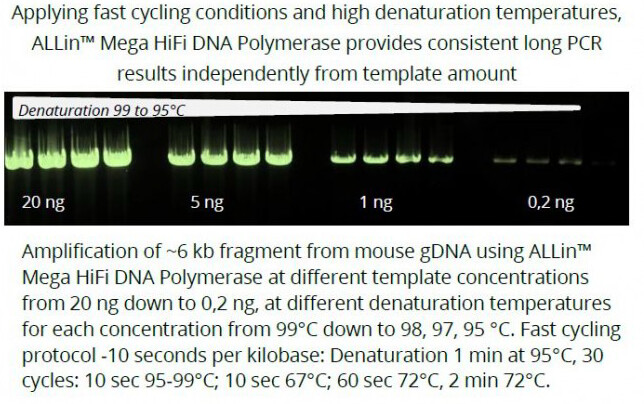
The new ALLin™ Mega HiFi DNA Polymerase is the perfect choice for NGS applications.
Fast high yield PCR with fidelity 100x higher than Taq. For amplification also of GC/AT-rich templates.
ALLin Mega HiFi is available in a range of formats:
Hotstart or nont-notstart polymerase
Separate polymerase with buffer including all necessary components
Mastmix with or without red dye

Hybridisation capture enrichment in targeted NGS, using a novel probe design technology, increases data uniformity compared to PCR amplification enrichment. Hybridisation capture gives coverage across UTR´s, Promoters, CDS, Splicing sites and enables detection of more mutants; insertions/deletions, CNV´s, SNV´s and gene fusion.
The unique re-balancing process with full QC will ensure optimal design and efficient workflows.
Download comparison data here!
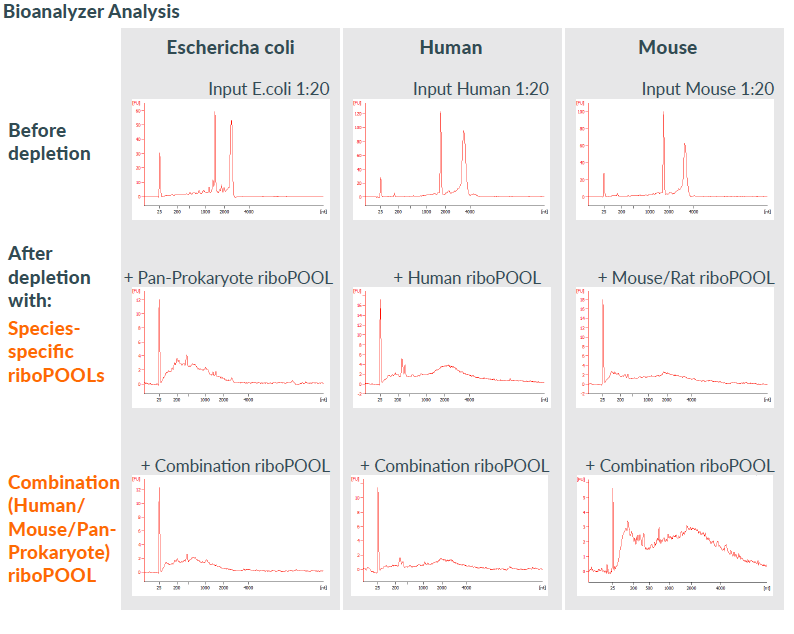
The riboPOOL™ offers simultaneous ribo-depletion for complex samples. The method will deplete ribosomal RNA simultaneously across multiple species.
Efficient depletion of human, mouse and E. coli RNA with the Combination riboPOOL consisting of a 1:1:1 mix of Human, Mouse/Rat and Pan-Prokaryote riboPOOL was observed for 1 μg RNA input.
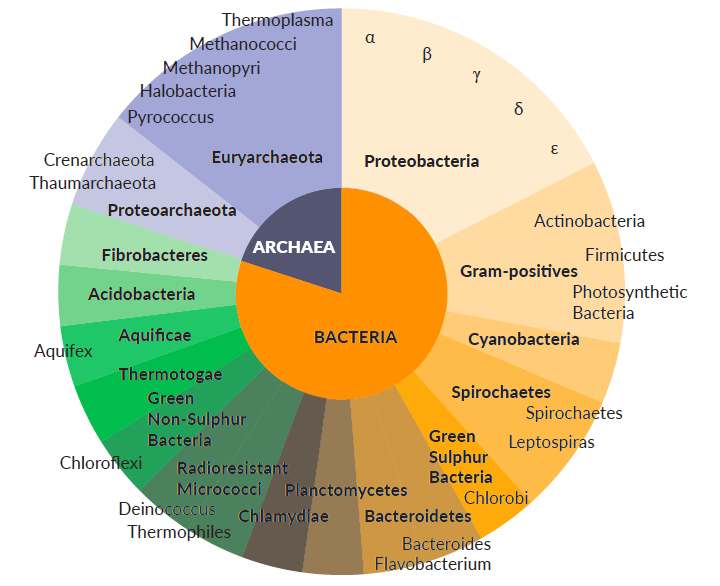
The Pan-Prokaryote riboPOOL gives the widest microbial coverage for microbiome/metagenomic analysis, covering >75 different species simultaneously. Removing ribosomal RNAs (rRNAs) with high efficiency and targets 5S, 16S and 23S rRNA.
For metagenomics, the riboPOOLs are available as combinational reagent, for example as Pan-prokaryote:Human mix, based on the expected ratios of your sample.
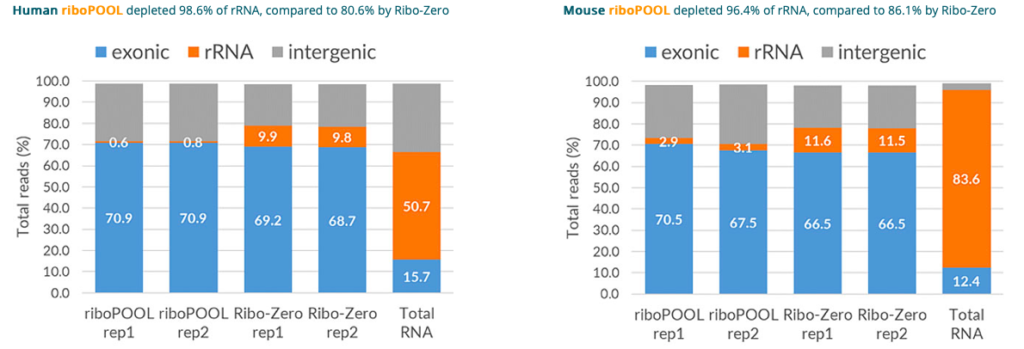
Removing ribosomal RNAs (rRNAs) to allow sensitive detection of scientifically relevant RNAs by Next Generation Sequencing, is currently performed with costly kits limited to well-studied species.
riboPOOLs present an affordable and flexible solution that gives scientists absolute freedom to deplete rRNAs or other custom RNAs from any species.
Composed of high complexity pools of optimally-designed biotinylated DNA probes, riboPOOLs even outperformed Ribo-Zero (Illumina), depleting ~10% more rRNA while leaving other RNA intact.
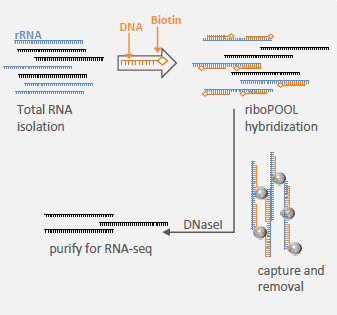
riboPOOLs are available for any species. Contact us for your specific needs.
riboPOOLs in stock: Human, Mouse, Planarian, Drosophila, Silkworm. Available in 2, 5 or 10 nmol (20, 50, 100 reactions). Custom scales on request.
Protocol is based on biotin-streptavidin chemistry with magnetic bead capture and removal.
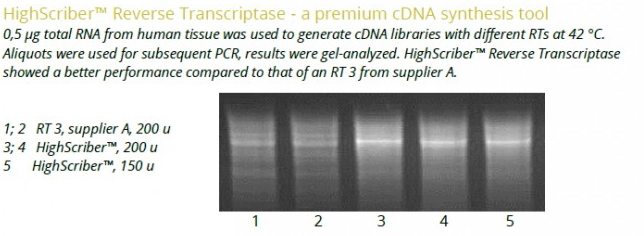

Thermostable Reverse Transcriptase blended with Ribonuclease Inhibitor for an efficient cDNA synthesis
High yields of full lengths transcripts up to 12-15 kb
cDNA synthesis from complex templates at up to 55°C
High sensitivity detection from 1 pg of total RNA template
Request your free sample: info@lablifenordic.com
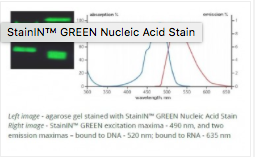
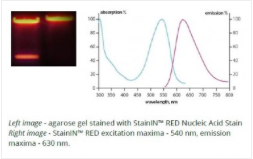

Twice as concentrated than other stains, make StainIN™ GREEN and StainIN™ RED Nucleic Acid Stains much more economical. You get highly sensitive, and non-toxic detection of gels. Developed by the professionals at HighQu.
Replace your green dye with StainIN™ GREEN:
2X more stain for the same price
Blue LED or UV detection compatibility
4X more sensitive to EtBr
Unique emission pattern stains RNA and DNA in different colours (RNA in red, DNA in green)
Replace your red dye with StainIN™ RED:
2x more concentrated than other red dyes
2X more sensitive than EtBr
Time saving staining of gels
Much more safe than EtBr
Ordering information:
NAS0201 StainIN™ GREEN Nucleic Acid Stain, 1 ml, 20000X; SEK 1.097.-; DKK 857.-
NAS0101 StainIN™ RED Nucleic Acid Stain, 1 ml, 20000X; SEK 1.097.-; DKK 857.-
Place your order here: info@lablifenordic.com
Product information sheet:
StainIN™ GREEN: StainIN GREEN Product Sheet
StainIN™ RED: StainIN RED Product Sheet
HighQu 2017 Catalogue: HighQu Catalogue 2017

The new mastermix is not only great for achieving best data. It also includes an inert blue dye for improved sample visibility during handling and pipetting. The highly experienced team at HighQu have developed a range of great performing PCR reagents.
The mastermixes give you
qPCR on instruments calibrated for low or high ROX concentration
qPCR from gDNA, cDNA, viral DNA, low copy number genes
Universal – both standard and fast cycling
Easy protocols – minimum optimization
Excellent for GC or AT rich templates
Highest sensitivity, rapid extension, early Ct values; based on the small molecular inhibitor technology Hot Start PCR
Supplied with PCR Water to guarantee the best performance.
Data sheets, ORA™ SEE qPCR Green Mix with Low or High ROX content:
QPD05_ORA_SEE_qPCR_Green_ROX_L_Mix_PI-1
QPD04_ORA_SEE_qPCR_Green_ROX_H_Mix_PI-1
Ready to test the ORA™ SEE qPCR Green Mix?
info@lablifenordic.com
Menu bar
Contact information
Follow us